Research News
Cell type- and Projection-specific Brain-wide Reconstruction of Single Neurons
On November 19th, 2018, Dr. Minmin Luo’s laboratory published a paper entitled “Cell type- and Projection-specific Brain-wide Reconstruction of Single Neurons” in Nature Methods. In this study, they described the development of a dual-AAV expression system that enables strong and sparse labeling of individual neurons with cell type- and projection-specificity. By combining with fMOST and other imaging techniques, this system can be used to achieve whole-brain reconstruction of individual neurons.
Mapping the neuronal connections between different brain areas has been a major theme of neuroscience research. In the past 5 years, there has been tremendous progress in the mapping of the connections of bulk-labeled neuron populations on the mesoscale level using light microscopies. Recent efforts have been more directed at studying the precise morphology of individual neurons at the whole-brain level. However, two technical challenges remain to be the hindrance of this progress: first, in order to clearly resolve the neuronal processes, neurons must be labelled sparsely and brightly; and second, in order to obtain accurate spatial context within the whole brain, the brain sample must be handled with a reliable imaging system of adequate resolution, and computational pipelines need to be devised to analyze the vast amount of data produced.
To address the first issue, researchers from Dr. Minmin Luo’s lab developed a dual-AAV expression system that sparsely and strongly labels neurons with cell-type specificity. The labeling system consists of two AAV vectors: the “Controller” and the “Amplifier”. The “Controller” governs the initiation of sparse expression, whereas the “Amplifier” enhances the signal once the labeling is initiated (Fig. 1). The researchers showed that with the aid of recombinase pairs, the labeling was restrained to specific molecular cell types. They also demonstrated that by tuning the mix ratio of “Controller” and “Amplifier”, the labeling sparseness can be fine-tuned.
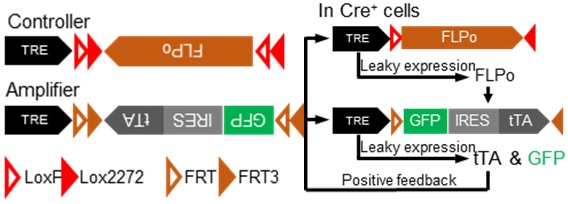
Figure 1. Working principles of the sparse labeling system
To resolve the second technical challenge, Dr. Minmin Luo’s lab established collaboration with the fMOST team from Wuhan National Laboratory for Optoelectronics, Huazhong University of Science and Technology (HUST), and devised a comprehensive pipeline including viral sparse labeling, specimen embedding, fMOST imaging, digital reconstruction, data registration and quantitative analysis. With the aid of this pipeline, the researchers achieve complete reconstruction of 15 dopamine neurons on the whole-brain level. The reconstructed dopamine neurons could be classified into two types based on their morphologies: one with restricted projection into single downstream targets, forming densely distributed arborizations at its terminal; the other with profuse projection towards multiple targets, forming relatively fewer arborizations (Fig. 2).


Figure 2. 15 completely reconstructed dopamine neurons revealing two projection patterns
By incorporating a special type of AAV serotype, AAV-retro, researchers extended the sparse labeling system and achieved projection-specific sparse labeling. Using different injection paradigms, the researchers validated this strategy by labeling and reconstructing two cortical-striatal projection neuron types: the intratelencephalic neurons, projecting to bilateral dorsal striatum, and the pyramidal tract neurons, projecting to unilateral dorsal striatum (Fig. 3).



Figure 3. Projection specific labeling and whole-brain reconstruction of IT and PT neurons
An exciting trend in the field of optical imaging is the development of tissue clearing techniques. Researchers from Dr. Minmin Luo’s lab combined the sparse labeling system with uDISCO clearing techniques to image and reconstruct single neurons in thick brain section samples. They labelled two types of molecularly defined striatal interneurons: PV+ and SOM+, and focused on their dendritic morphologies (Fig. 4). Complete dendritic morphologies of PV and SOM neurons were rapidly reconstructed and analyzed, demonstrating that the labeling system is flexible and easily compatible with traditional imaging platforms.


Figure 4. Rapid reconstruction of PV and SOM interneurons achieved by sparse labeling and tissue clearing
In the initial design of the sparse labeling viruses, the researchers used Cre/FLP as the recombinase pair. Researchers introduced another pair of recombinases DreO/vCre and developed a second version of the sparse labeling system (Fig. 5 left). Multi-color labeling in the same mouse brain was achieved using these two versions of sparse labeling system, which demonstrated the generality of the dual-AAV strategy (Fig. 5 right).


Figure 5. Introducing additional recombinase pairs to speed up labeling efficiency
The sparse labeling system described here is flexible, efficient and easy to use. Besides labeling neurons for morphological studies, our approach can serve as a general sparse expression strategy to deliver various genetically-encoded effectors into specific neuron types to achieve optogenetic manipulation, imaging, electrophysiology, and gene-editing at the single-neuron level. The flexibility of this labeling system also lays the ground for its future application in various imaging techniques such as light-sheet microscopy. The development of this labeling strategy shall extend our ability to resolve the brain connectivity at individual neuron level and contribute to the study of single neuron morphology under normal and pathological conditions.
Dr. Rui Lin from Dr. Minmin Luo’s lab, Ruiyu Wang from Dr. Minmin Luo’s lab, and Dr. Jing Yuan from the fMOST team in HUST are co-first authors of this paper. Dr. Minmin Luo and Prof. Hui Gong from the fMOST team are co-corresponding authors. Other contributing authors include Qiru Feng and Youtong Zhou from Dr. Minmin Luo’s lab; Dr. Shaoqun Zeng and Dr. Miao Ren, Siqi Jiang, Hong Ni and Can Zhou from the fMOST team. This study was supported by MOST 973 grants, NNSFC, and Beijing Municipal Government. All works were completed in NIBS and Britton Chance Center for Biomedical Photonics, Wuhan National Laboratory for Optoelectronics - HUST.



