Research News
Dr. Li-Lin Du’s laboratory revealed the molecular basis of the non-autophagy function of Atg8
On November 19, 2018, Dr. Li-Lin Du's laboratory published in eLife a paper titled "Lipidation-independent vacuolar functions of Atg8 rely on its noncanonical interaction with a vacuole membrane protein". This study uncovered the molecular basis of the enigmatic non-autophagy function of the core autophagy protein Atg8.
Atg8 is a ubiquitin-like protein (UBL) that plays a central role in autophagy. Unlike other UBLs, Atg8 is conjugated to a lipid substrate, phosphatidylethanolamine, in a process termed lipidation. The lipidation of Atg8 is essential for autophagy in most of the systems where autophagy has been studied. Thus, it came as surprises when lipidation-independent roles of Atg8 were discovered, first in yeasts, and then in animals. Molecular mechanisms underlying the lipidation-independent functions of Atg8 have remained elusive.
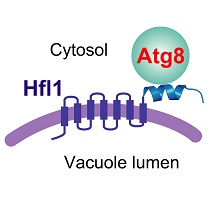
In this paper, the authors provide insights into this puzzle by identifying an Atg8-binding protein required for the lipidation-independent functions of Atg8 in maintaining normal vacuole morphology in two divergent model yeast species, the fission yeast Schizosaccharomyces pombe and the budding yeast Saccharomyces cerevisiae. This Atg8-binding protein, Hfl1, is a conserved 7-transmembrane protein related at the sequence level to both transporters and GPCRs. Its ability to recruit Atg8 to the membrane provides a satisfactory explanation on how Atg8 can function in membrane-related processes without lipidation.
Remarkably, through structural biology analyses of the Atg8-Hfl1 complexes, the authors discovered a completely new mode of Atg8 binding. The Atg8-binding regions of Hf1l proteins adopt a helical conformation, rather than the extended β strand conformation of conventional Atg8-interacting motifs. The surprising noncanonical way of Atg8 binding by Hfl1 proteins expands our understanding of the structural diversity of Atg8 binding modes.
This work is mainly a collaboration between Dr. Li-Lin Du’s lab at NIBS and Dr. Nobuo N. Noda’s lab at Institute of Microbial Chemistry in Japan. Xiao-Man Liu and Xiao-Min Du of the Du lab and Akinori Yamasaki of the Noda lab are co-first authors of this paper. Other contributors include Valerie C. Coffman and Dr. Jian-Qiu Wu of Ohio State University, Dr. Yoshinori Ohsumi and Dr. Hitoshi Nakatogawa of Tokyo Institute of Technology. Dr. Nobuo N. Noda and Dr. Li-Lin Du are co-corresponding authors. This project was supported by the Chinese Ministry of Science and Technology and the Beijing municipal government.
Web address of the paper:https://elifesciences.org/articles/41237



