Research News
Studies from Dr. Feng Shao's laboratory reveal the molecular mechanism of blocking host death receptor signaling by bacterial-effector-catalyzed arginine GlcNAcylation
On April 10, 2019 ---- Studies from Dr. Feng Shao's laboratory reveal the molecular mechanism of blocking host death receptor signaling by Enteropathogenic E. coli (EPEC) arginine GlcNAcyltransferase NleB. The work entitled "Structural and functional insights into host death domains inactivation by the bacterial arginine GlcNAcyltransferase effector" is published in the journal Molecular Cell.
Back in 2013, Dr. Shao reported in Nature that EPEC employs the type III secretion system (TTSS) effector NleB to block host death receptor signaling and facilitate bacterial survival and colonization in the mouse colon. Mechanistically, NleB catalyzes the attachment of N-acetylglucosamine (GlcNAc) onto a crucial arginine of the death domain (DD) and thereby disrupts DD interactions and formation of death receptor signaling complexes. NleB-catalyzed arginine GlcNAcylation is different from known glycosylation modifications and represents a novel type posttranslational modification (PTM). However, the precise mechanism underlying NleB recognition of DD substrates and catalyzing arginine GlcNAcylation has been unknown.

Figure 1. Crystal Structures of NleB and NleB-GlcNAcylated Death Domains
In this study, researchers from Dr. Feng Shao’s laboratory first determined high-resolution crystal structures of arginine-GlcNAcylated DD of TRADD and RIPK1, which demonstrates the GlcNAc is covalently linked to the Nε atom of arginine guanidinium via a β-anomeric configuration. They further performed NMR analyses and confirmed the β-anomeric configuration in the solution state of GlcNAcylated TRADD-DD and RIPK1-DD. The researchers also determined crystal structures of NleB alone and in complex with FADD-DD, UDP, and Mn2+. The structure of NleB bears an extended Rossmann-like fold, typical of the GT-A-type glycosyltransferases, and an antiparallel helix-pair insertion, which constitutes a “C”-like shape. The globular FADD-DD is elegantly accommodated by the large concave surface of NleB through extensive hydrogen-bond and electrostatic interactions between the enzyme and the substrate. NleB-modified arginine is highly conserved in 12 of the 34 human DD proteins, among which the adaptors TRADD, RIPK1 and FADD, and the receptors TNFR1, FAS, DR3, DR4 and DR5 function in death receptor signaling. Profiling the modification by co-expression with NleB and mass spectrometry analyses revealed DD of TRADD, RIPK1, FADD, TNFR1, FAS, and DR3, but not those of DR4 and DR5, were GlcNAcylated by NleB. Sequence alignment and structural comparison showed that the charged residues mediating the interaction of DD with NleB were highly conserved in NleB-sensitive DD. By contrast, most of these charged residues involved in NleB recognition were substituted by oppositely charged residues in NleB-resistant DR4- and DR5-DD. Mutations that reversed the charge state were introduced into DR5-DD or NleB, which enables NleB to GlcNAcylate mutated DR5-DD or render wildtype DR5-DD GlcNAcylation by engineered NleB. The enzyme-substrate complex structure also reveals the interactions of sugar donor UDP-GlcNAc, the co-factor Mn2+, and the acceptor arginine in the catalytic pocket of NleB. The geometry of the sugar donor and the position of the arginine acceptor reveals a complete mechanism of NleB-catalyzed sugar transfer: the side chain of acceptor arginine stretches into a narrow cleft in NleB, generating a bidentate hydrogen-bond interaction between the arginine guanidinium and the carboxyl group of Glu253; Glu253 of NleB acts as the base to deprotonate the guanidinium in the arginine substrate; the arginine then nucleophilically attacks the C1 atom of UDP-GlcNAc, forming an oxocarbenium ion-like transition state that progresses to SN2-like displacement of the leaving group UDP. These mean that NleB employs an inverting sugar-transfer mechanism to convert α-anomeric configuration of C1 in UDP-GlcNAc into a β-glycosidic linkage. The researchers also performed mutagenesis of the substrate recognition interface and the catalytic pocket of NleB to verify the structural basis of NleB GlcNAcylation of host DD proteins by in vitro GlcNAcylation and NleB-mediated blocking of host cell death, and then performed mouse infection assays to investigate the functional significance of above structural findings. Moreover, by co-immunoprecipitation assays, they found NleB is a broad-spectrum inhibitor of death receptor signaling pathways due to its pleotropic blocking of DD interactions.
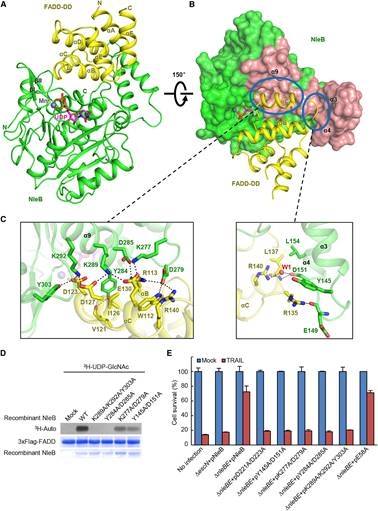
Figure 2. Crystal Structure of the NleB/FADD-DD Complex and Analyses of the Binding Interface
This study expands our knowledge about bacterial effectors manipulation of host signaling pathways via novel PTMs, and shall guide the NleB-based drug development for antagonizing EPEC infection. More importantly, NleB-catalyzed arginine GlcNAcylation is different from other known glycosylation, and the findings reported in this work provide a comprehensive and in-depth understanding of the enzymatic reaction mechanism of arginine GlcNAcylation. NleB and NleB-like activity may also serve as a useful tool for dissecting the complicated functional and regulatory mechanisms of host death receptor signaling.
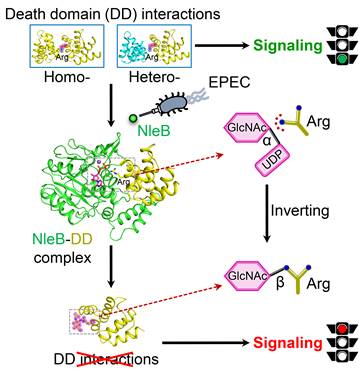
Figure 3. A structural model of NleB modification and inactivation of host death-domain signaling pathway.
Visiting scientist Dr. Jingjin Ding from Institute of Biophysics (CAS) is the first author of this paper. Other contributors include graduate students from Dr. Shan Li’s group (a former PhD student of Dr. Shao, now a PI at Huazhong Agricultural University), Dr. Qin Yao (a former PhD student of Dr. Shao, now a postdoc fellow at Caltech), Dr. Hongwei Yao from the High-Field Nuclear Magnetic Resonance Center at Xiamen University, and Prof. Da-Cheng Wang form Institute of Biophysics (CAS). Drs. Jingjin Ding, Shan Li, and Feng Shao are the co-corresponding authors. The study was supported by the National Key Research and Development Programs of China, the Strategic Priority Research Program of CAS, the program Youth Innovation Promotion Association of CAS, National Natural Science Foundation of China, and carried out at National Institute of Biological Sciences, Beijing.



